Wrapper script
library(dynwrap)
library(dyncli)An alternative to wrapping a script inside R, is to wrap it using an external script. Because this does not provide any dependency management, this is not really useful for method end-users, but rather as a way to easily develop a TI method and to ultimately step up towards containerised wrapping.
Similarly as a wrapper written in R, you’ll need to provide both a definition (= a definition.yml) and a way to run the methods (= a script).
definition.yml
The definition has the same hierarchical structure as used by the definition() function. A minimal example
method:
id: comp_1
parameters:
- id: component
default: 1
type: integer
distribution:
type: uniform
lower: 1
upper: 10
description: The nth component to use
wrapper:
input_required: expression
input_optional: start_id
You can use our template definition.yml to see all the different possibilities of what can be included in the definition: https://github.com/dynverse/dynmethods/tree/master/template_container . It may also be useful to check out some of the wrappers in dynmethods, e.g.: https://github.com/dynverse/ti_paga/blob/master/definition.yml
Running script
A running script reads in the data, creates a trajectory, and again writes the trajectory to a file. To do the loading and writing tasks, we provide helper libraries for R and python: dyncli and dynclipy.
The wrapper script will typically have the following structure:
- A shebang, such as
#!/usr/bin/env Rscriptor#!/usr/bin/env python - A call to dyncli(py) to load in the data, such as
dataset <- dyncli::main()ordataset = dynclipy.main() - All you need to infer the trajectory
- Constructing the trajectory model using any of the wrapping methods
- A call to dyncli(py) to write the data, such as
dyncli::write_output(trajectory)ortrajectory.write_output()
A minimal example script for R:
#!/usr/bin/env Rscript
dataset <- dyncli::main()
library(dynwrap)
library(dplyr)
# infer trajectory
pca <- prcomp(dataset$expression)
pseudotime <- pca$x[, dataset$parameters$component]
# flip pseudotimes using start_id
if (!is.null(dataset$priors$start_id)) {
if(mean(pseudotime[start_id]) > 0.5) {
pseudotime <- 1-pseudotime
}
}
# build trajectory
trajectory <- wrap_data(cell_ids = rownames(dataset$expression)) %>%
add_linear_trajectory(pseudotime = pseudotime)
# save output
dyncli::write_output(trajectory, dataset$output)
Make sure this script is executable!
chmod +x run.Rand for Python:
#!/usr/bin/env python
import dynclipy
dataset = dynclipy.main()
import pandas as pd
import sklearn.decomposition
# infer trajectory
pca = sklearn.decomposition.PCA()
dimred = pca.fit_transform(dataset['expression'])
pseudotime = pd.Series(
dimred[:, dataset['parameters']['component']-1],
index = dataset['expression'].index
)
# build trajectory
trajectory = dynclipy.wrap_data(cell_ids = dataset['expression'].index)
trajectory.add_linear_trajectory(pseudotime = pseudotime)
# save output
trajectory.write_output(dataset['output'])
Testing it out
method <- create_ti_method_definition(
"definition.yml",
"run.R"
)
dataset <- dynwrap::example_dataset
trajectory <- infer_trajectory(dataset, method(), verbose = TRUE)## Executing 'comp_1' on 'example'
## With parameters: list(component = 1L),
## inputs: expression, and
## priors :
## Input saved to /tmp/RtmpwQdLKe/file68d26110ae05/ti
## Running ./run.R --dataset input.h5 --output output.h5
##
## Attaching package: ‘dplyr’
##
## The following objects are masked from ‘package:stats’:
##
## filter, lag
##
## The following objects are masked from ‘package:base’:
##
## intersect, setdiff, setequal, union# install dynplot to plot the output
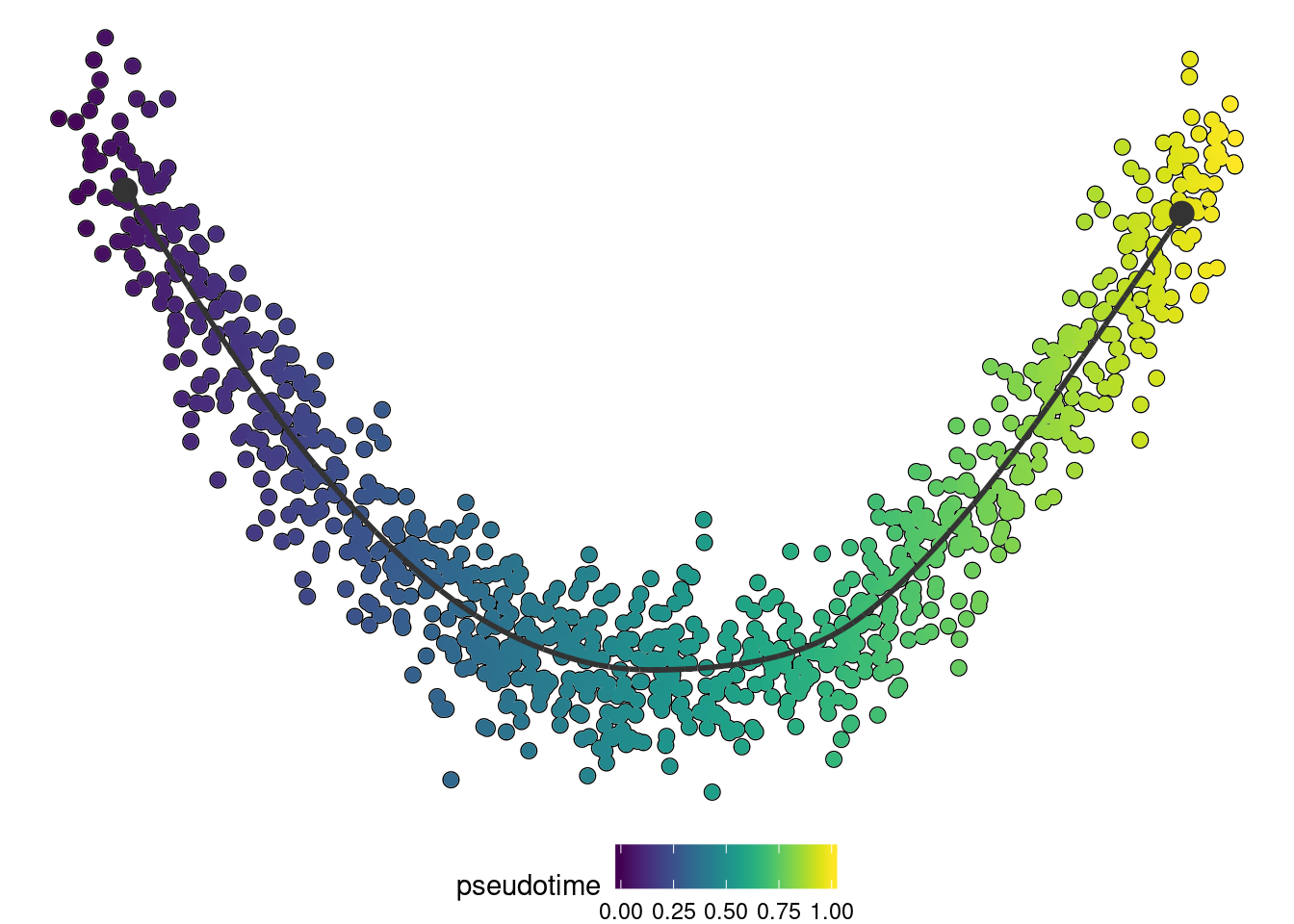
if ("dynplot" %in% rownames(installed.packages())) {
dynplot::plot_dimred(trajectory, color_cells = "pseudotime" , expression_source = as.matrix(dataset$expression))
}
Interactive development
With the debug parameter, it is possible to enter the script interactively. This makes it easier to develop TI method as you can just load in the input data, and code!
infer_trajectory(dataset, method(), debug = TRUE)Making your method available for other users
Wrapping a method inside a script does not have any dependency management, and is therefore only meant for development purposes. To deploy your method to other users, check out the containerisation tutorial!
Wrapping a method without dyncli(py)
While using dyncli to load and save the data is the most straightforward when using R and python, it’s also possible to skip these packages and do all the work yourself. Briefly you have to:
- Parse the arguments provided by dynwrap yourself, e.g.
--dataset input.h5. - Read in the hdf5 files generated by dynwrap
- Write the output file in the correct hdf5 format
If after this you’re still convinced you want to do this, please contact us. We’ll be able to provide some further documentation!